Quantitative STEM Analysis of Supramolecular Assemblies
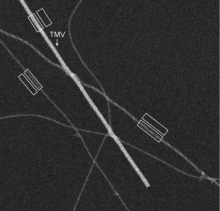
The purpose of this research is to develop techniques for determining quantitative structural information from supramolecular assemblies using the technique of scanning transmission electron microscopy (STEM). In STEM a finely focused probe of high-energy electrons is scanned across a biological structure of interest and various signals are detected at each image pixel. For example, we are using the annular dark-field signal to determine the molecular masses of large protein assemblies, e.g., Alzheimer’s disease-related amyloid fibrils; such measurements provide information about arrangement of subunits.
Novel Approaches for Determining 3D Cellular Ultrastructure
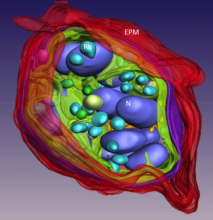
We are also developing techniques for determining subcellular structure based on axial bright-field STEM tomography by recording images from micrometer-thick sections of cells over a range of tilt angles. Using this approach, we have shown that it is possible to reconstruct much thicker volumes than can be achieved with conventional TEM electron tomography. We are currently applying STEM tomography to visualize entire synapses in the nervous system at a spatial resolution of a few nanometers. Of particular current interest is to determine the structure of postsynaptic densities in hippocampal neurons, as well as ribbon synapses in rod bipolar cells of retina. We are also using STEM tomography to study a number of biological systems outside the field of neurobiology, including the structural changes that occur on activation of human blood platelets.
Quantitative Nanoscale Analysis using Inelastic Electron Scattering
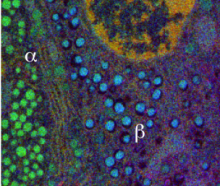
In this research, we are developing techniques to map chemical elements contained in subcellular structures using (1) energy-filtered transmission electron microscopy (EFTEM), and (2) STEM coupled with electron energy loss spectroscopy (EELS). In both approaches, we acquire hyperspectral inelastic images carrying information from characteristic excitations of atomic core-shells in the specimen. Our aim is to extract quantitative composition and to achieve a sensitivity of a few atoms. Recent applications include imaging the iron cores of individual ferritin molecules in neurons and other cells, where iron plays an important physiological role. In another application, we are combining EFTEM with electron tomography to map DNA and protein in three-dimensions within cell nuclei by using the phosphorus and nitrogen signals.
Methods for 3D Reconstruction
Tomographic reconstructions based on the standard weighted back-projection (WBP) algorithm suffer from artifacts due to the limited available angular tilt range. Algorithms such as the simultaneous iterative reconstruction technique (SIRT), which we have implemented in our laboratory, provide some improvements in the quality of 3D reconstructions but artifacts still persist. We are exploring other techniques by introducing some prior knowledge about the reconstructed volume by the application of regularization conditions.
Characterization of Bionanoparticles
Bionanoparticles being developed for potential use in theranostic nanomedicine often have a hybrid structure that incorporates both organic and inorganic components. STEM and EFTEM provide unique information about the arrangement and proportions of the chemical constituents that are critical in controlling the function of such hybrid bionanoparticles. We are using these methods to characterize bionanoparticles containing a variety of moieties including MRI contrast agents, optical fluorescence probes, nanogold atomic clusters, carbon nanotubes carriers, as well as anti-cancer drugs such as doxorubicin.